Norovirus (NoV) is responsible for 30%–80% of acute gastroenteritis (AGE) outbreaks in long-term care facilities (LTCFs) and hospitals in developed countries.Reference Kambhampati, Koopmans and Lopman 1 , Reference Chen, Hall, Kirk, Health and Diseases 2 The mainstay of NoV outbreak management is an effective infection prevention and control program because the treatment of NoV is supportive care.Reference Robilotti, Deresinski and Pinsky 3 Multiple routes of transmission, high infectiousness, and environmental stability of NoV make the control of NoV outbreaks complex and difficult.Reference Kambhampati, Koopmans and Lopman 1 Additionally, NoV are usually underdiagnosed in healthcare institutions, which has implications not only for costs but also clinical impact. Noroviruses cause particularly severe disease in elderly patients and in people with pre-existing conditions.Reference Beersma, Sukhrie and Bogerman 4 , Reference Cardemil, Parashar and Hall 5
Genogroups I (GI), II (GII), and IV (GIV) are the 3 groups known to cause diarrheal disease in humans. GII is the most commonly associated with norovirus infection, and since 2002, GII genotype 4 (GII.4) has been recognized as the major cause of AGE outbreaks occurring worldwide.Reference Capece and Gignac 6 Due to evolutionary mechanisms of recombination and mutation, new GII.4 variants emerge every 2–4 years.Reference Parra, Squires and Karangwa 7 More recently, non–GII.4 strains, including GII.17 and GII.2, have been the main epidemic strains, together with GII.4 strains, in several countries.Reference da Silva Ribeiro de Andrade, Fumian and Leite 8 – Reference Kim, Song and Lee 11 Between October 31 and December 8, 2017, an outbreak of AGE caused by NoV occurred in Mafra, Portugal, in a long-term care facility (LTCF). We investigated the outbreak to describe and estimate its extent, and we implemented control measures.
Methods
Outbreak setting
The LTCF where the outbreak occurred is a certified institution for continuous care. The LTCF includes several services and departments for residents and outpatients: long-term care department, healthcare and rehabilitation, outpatient clinic, and social center. Communal facilities include gymnasium for rehabilitation, therapeutic pool, social area for residents and visitors, and a dining room. The LTCF has a total of 11 wards with 335 residents in different typologies: 60 patients who came from central hospitals for supportive medical and nurse care; 200 patients from the National Network for Integrated Continuous Care, and the rest were residents in a particular regimen (eg, a retirement home). The wards are distributed over 4 floors, and each room is occupied by at least 3 residents. Residents were aged 47–96 years old and were mostly female. Staff members include doctors, nurses, cleaning, laundry and cooking staff, and other staff members such as pharmacists, physiotherapists, psychologists, and social workers. The professionals do their work in all wards and contact with all autonomous residents and staff in the common areas of the building.
Case definition
We defined probable cases as residents or staff members in the LTCF with at least 1 of the following symptoms between October 31 and December 8, 2017: (1) diarrhea (watery or soft stool) 3 or more times within 24 hours, (2) vomiting, (3) nausea, and/or (4) abdominal pain. Confirmed cases were probable cases whose stool specimen tested positive for NoV by real-time polymerase chain reaction (RT- PCR) and had the same genotype.
Epidemiological data collection
The nurses collected information from residents (included demented) and staff members identified as probable cases using a questionnaire on the day of onset of symptoms. Data included demographic information, signs and symptoms, date of onset, wards of residency (residents) or work (staff members), and history of contact with a person presenting with diarrhea or/and vomit. Retrospectively, staff members filled epidemiological questionnaires to validate data collected during the outbreak. We calculated incidence rates with 95% confidence interval (CI) as the number of probable cases of residents or staff members, divided by the total number of residents or staff members, respectively, in the LTCF during the outbreak period.
Microbiological investigation
Stool specimens were submitted for molecular detection of gastrointestinal virus, and 2 samples were also cultured for common enteropathogenic bacteria. Viral nucleic acid extraction from stools specimens was performed using the automated acid extraction platform EasyMAG (bioMérieux, Marcy-L’Étoile, France). Molecular detection was performed using a commercial multiplex RT-PCR assay for viral gastroenteritis pathogens (FTD Viral Gastroenteritis, Fast Track Diagnostics, Luxembourg) according to the manufacturer’s instructions. Genotype characterization of positive specimens was further performed using conventional PCR and sequencing analysis as previously described by Noronet network.Reference Vinjé, Hamidjaja and Sobsey 12 , Reference Fonager, Barzinci and Fischer 13 Nucleotide sequences of nonstructural polyprotein (ORF1) and capsid protein (ORF2) genes retrieved in this outbreak were deposited in GenBank under accession number MH930953. Genotype assignment was performed using the alignment from a public automated genotyping tool from Noronet platform (http://www.rivm.nl/en/Topics/N/Noronet/Databases/Sequence_typing_tool) and other retrieved sequences from GenBank.Reference Kroneman, Vennema and Deforche 14 Phylogenetic trees (based on the partial sequences of ORFs 1 and 2) were constructed using the neighbor-joining method with 1,000 bootstrap replicates (ie, a Kimura 2-parameter model) in MEGA version 7.0 software (Pennsylvania State University, University Park, PA).Reference Kumar, Stecher and Tamura 15
Results
Outbreak description
The main responsible members of the LTCF observed the outbreak began on November 2. However, on November 3, the number of cases suddenly dropped and the outbreak was thought to have ended. In the following days, cases increased again, and the outbreak was reported to the local public health unit on November 8. We identified 146 cases among 97 residents and 49 staff members, with an incidence rate of 29% (97 of 335; 95% confidence interval [CI], 24.2–34.1) among residents and 13.9% (49 of 352; 95% CI, 10.5–18) among staff. The incidence rates among medical staff were 19.3% (16 of 83; 95% CI, 11.4–29.4) among nurses and 9.1% (8 of 88; 95% CI, 4–17.1) among other 8 staff members. In addition, the incidence rate was 13.8% (25 of 181; 95% CI, 9.1–19.7) in the group of cleaning, laundry, and cooking staff members (Table 1). None of the staff members presented symptoms until October 31, 2017, or reported previous contact with cases of gastroenteritis. The median age among residents was 80 years (range, 47–96); ages of staff members were not available. The main symptoms among cases included diarrhea (121 of 146, 83%), vomiting (107 of 146, 73%), and to a lesser extent, nausea (13 of 146, 8.9%), fever (2 of 146, 1.4%), and abdominal pain (1 of 146, 0.7%). The index case was identified as a lucid 80-year-old female resident with onset of symptoms on October 31 (Fig. 1). She was transferred from a central hospital the day before the onset of symptoms but there was no information on AGE cases from that hospital. The outbreak affected all 11 wards and peaked 4 times on the following dates: November 2, 9, and 23, and December 2 (Fig. 2). On December 8, the final NoV cases were detected among residents in 3 different wards.
Table 1. Descriptive Analysis of the Outbreak Among Residents and Staff Members in a Long-Term Care Facility in Portugal, 2017 (n=146)
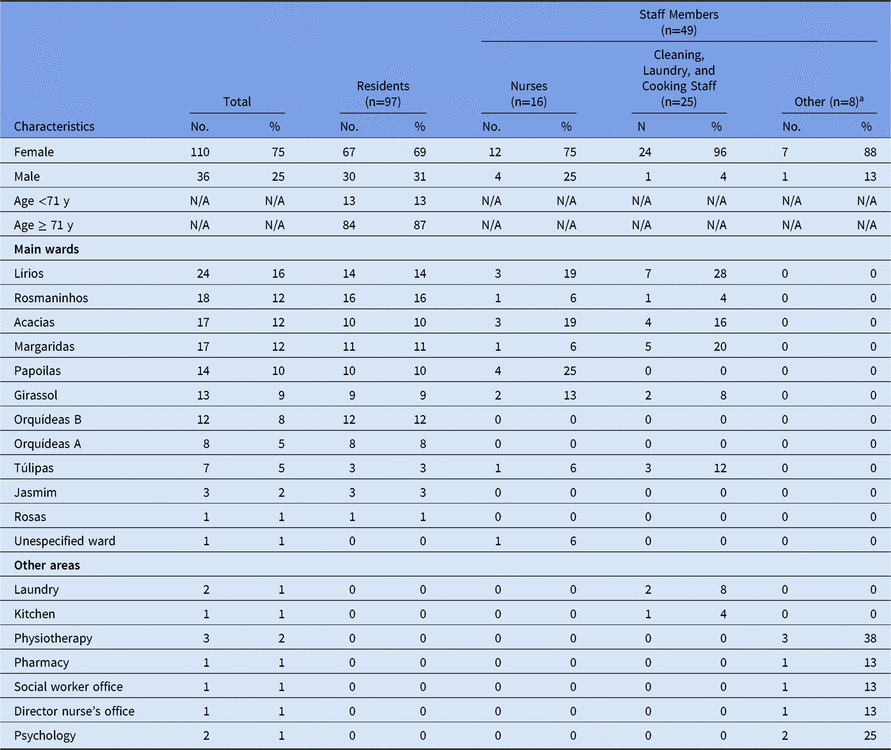
a Includes pharmacists, physiotherapists, psychologists, social workers and the director nurse.
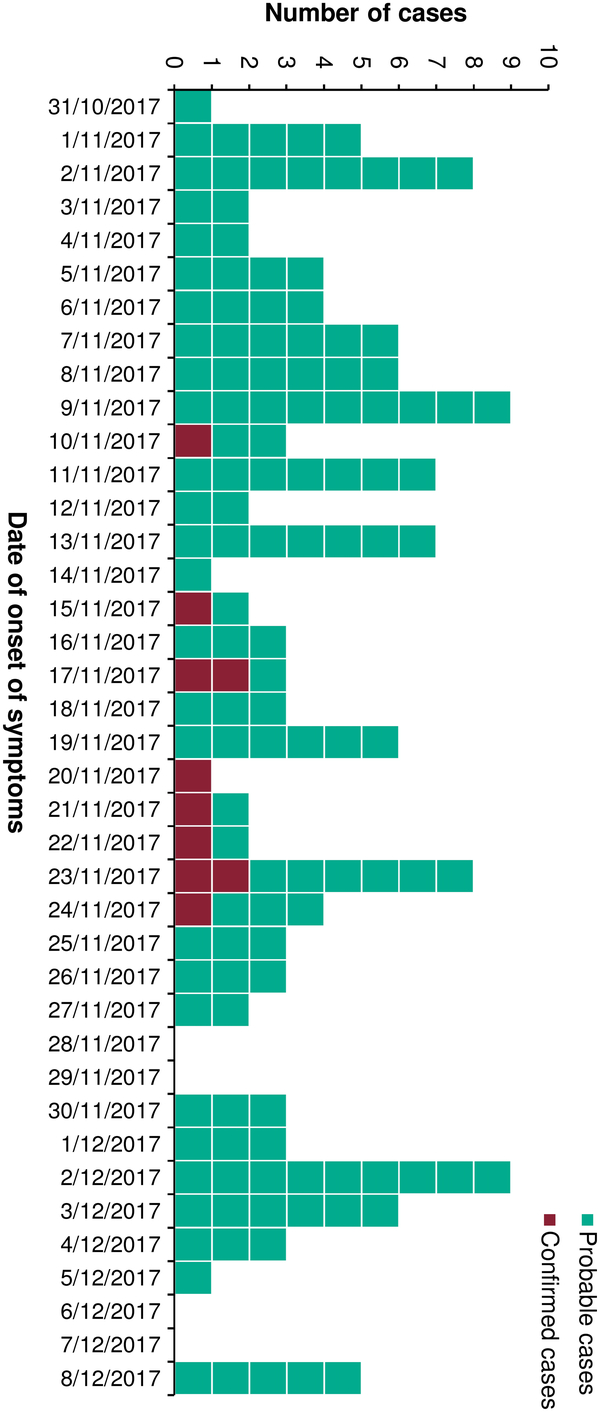
Fig. 1. Number of probable and confirmed norovirus cases, by date of onset of symptoms, during an acute gastroenteritis outbreak in a long-term care facility in Portugal in 2017 (n = 146).
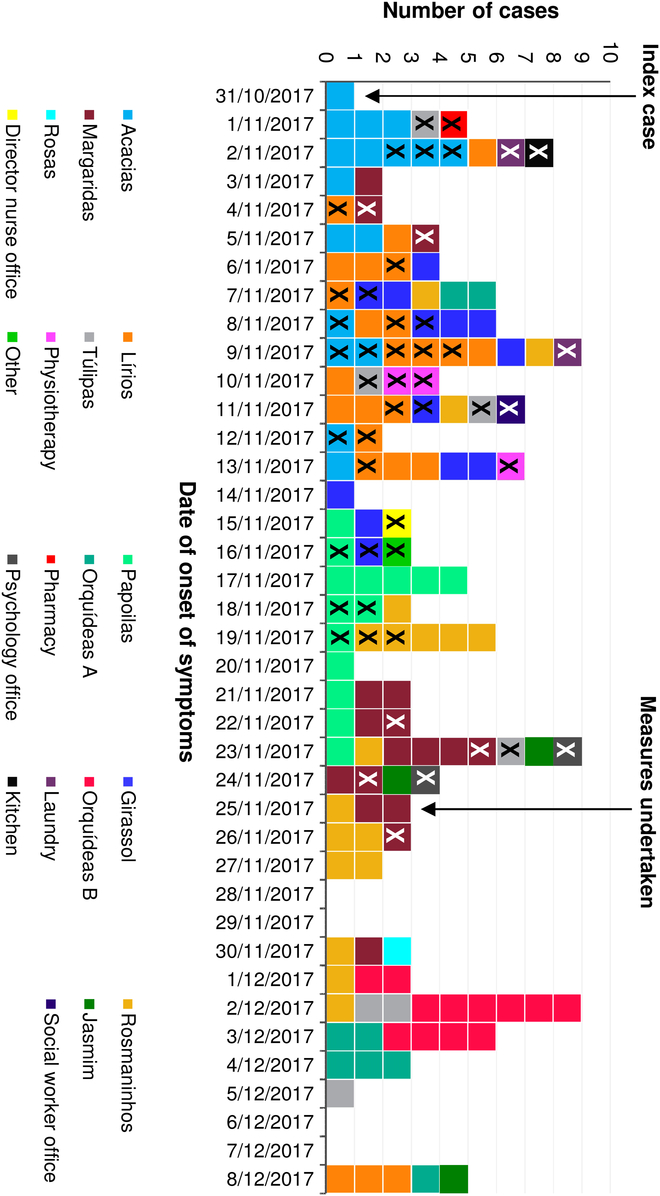
Fig. 2. Number of cases, by date of onset of symptoms and areas where cases stayed (residents, n=97) or worked (staff members, n = 49), during an acute gastroenteritis outbreak in a long-term care facility in Portugal in 2017. Square with an X indicates a staff member.
Laboratory tests, genotype determination, and phylogenetic analysis of NoV
On November 24 and 25, 10 stool samples were collected. All 10 samples were positive for NoV GII; however, in the 2 cultured stools, no bacterial pathogens were detected. All confirmed cases were residents whose onset of symptoms occurred on different days. A recombination of GII.P16 (ORF1) and GII.2 (ORF2), with a subclustering of Sydney 2012 (Fig. 3a and 3b) without nucleotide differences (or without single-nucleotide polymorphisms) was found in the stools of all confirmed cases.
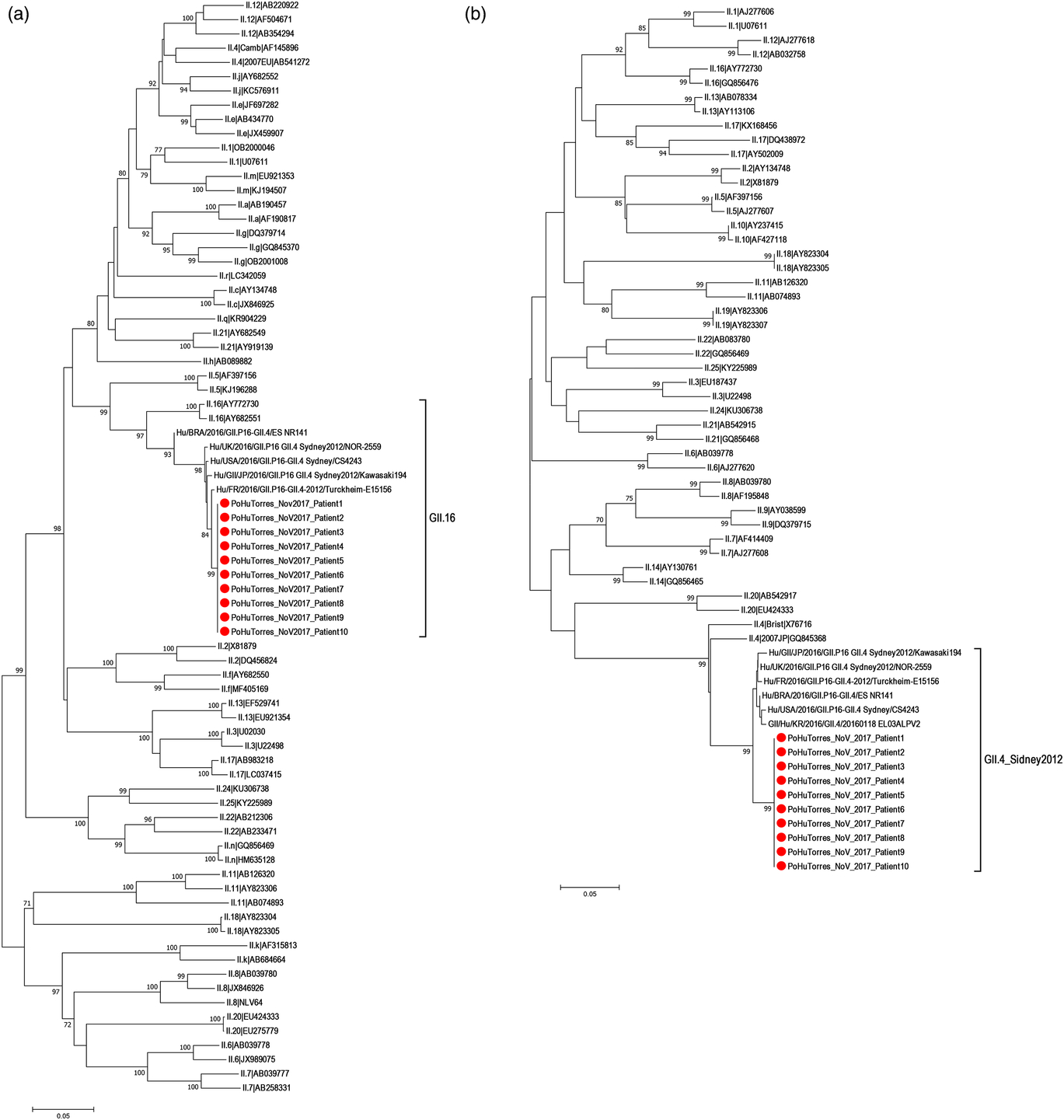
Fig. 3. Phylogenetic analysis of norovirus GII based on the partial nucleotide sequences of the nonstructural polyprotein (ORF1) and capsid (ORF2) regions: (a) based on an ORF1 745-bp fragment and (b) based on an ORF2 278-bp fragment. Phylogenetic trees were constructed using the neighbor-joining method with 1,000 bootstrap replicates (Kimura 2-parameter model) using MEGA version 7.0 software. Reference sequences for the alignment were retrieved from the typing tool from Noronet and GenBank. Only bootstrap values >70 were represented at the branch nodes. The GenBank accession number for the norovirus outbreak strain PoHuTorres_NoV_2017 sequence is MH930953. The red circle indicates outbreak strain sequences.
Outbreak control measures
NoV was suspected to be the causative pathogen of the outbreak based on Kaplan’s clinical and epidemiological criteria before the microbiological confirmation: vomiting in >50% cases; duration of illness 12–60 hours, incubation period 15–36 hours; and no presence of bacterial pathogens.Reference Kaplan, Feldman, Campbell, Lookabaugh and Gary 16 , Reference Turcios, Widdowson, Sulka, Mead and Glass 17 Based on these criteria, 1 week following the beginning of the outbreak, the LTCF staff implemented control measures including environmental disinfection, hand hygiene education, and use of personal protective equipment (PPE) to break transmission chains and control the outbreak. The outbreak was reported to the local public health unit on November 8, 2017. Adherence to the implemented control measures was insufficient due to staffing shortage. Therefore, on November 25, the local public health unit provided the LTCF with a written list of control measures to be rigorously implemented (Box 1). These upgrades to the control measures were based on the appropriate laundry handling and instructions to cleaning staff about the proper way to disinfect surfaces. The isolation and cohorting of cases was reinforced at that time to minimize resident and staff movements. There were no new admissions on the wards when a case was detected and until 2–3 days after total recovery of the last patient in that ward. The movement of staff members, including doctors, from wards where there were infected cases to wards where there were no cases was restricted. No further cases occurred among staff members after November 26 following implementation of these control measures.
Box 1. Measures Recommended by the Local Public Health Unit to Manage the Norovirus Outbreak by the Local Public Health Unit.
1. Emphasize the need for good hand hygiene practices using water and disinfectant soap for at least 20 seconds in staff members and residents.
2. Emphasize the need for good hand hygiene practices in visitors before and after contact with residents. Information about the preventive measures related to the hand hygiene was explained using posters at the entrance of the building.
3. Make the personal protection equipment (PPE, ie, lab coat, face mask, and gloves) available and provide clear information and supervision on its correct use for persons entering the patient care areas or caring for ill residents.
4. Intensify the environmental disinfection of surfaces with sodium hypochlorite. Moreover, clean and sterilize twice daily the room and surfaces in rooms where symptomatic residents stayed.
5. Isolate infected residents. The isolation must start with the onset of symptoms and last at least 48–72 hours after total recovery.
6. Staff members who present symptoms must stay at home for 48–72 hours after total recovery.
7. Minimize resident transfer between rooms and wards. Residents who present with symptoms must not be transferred between wards, and asymptomatic people must not be admitted in wards where symptomatic people reside.
8. Minimize the movement of staff members between wards with symptomatic and asymptomatic residents as much as possible.
9. Staff members must use proper PPE to manipulate all the dirty clothes, bed linen, and textiles with vomit or feces as well as for residents’ hygiene and rooms and sanitary facilities cleaning.
10. Staff members from laundry must manipulate all dirty textiles with gloves and, adequate disinfection cycles must be used for dirty textiles coming from wards with symptomatic residents.
11. Exclusion of ill food handlers until the Services of Health and Safety at Work has considered them suitable for their functions.
Discussion
We have described the first reported NoV outbreak in a LTCF in the country, which affected 146 people in an LTCF in Mafra, Portugal, caused by the emergent NoV GII.P16 GII.4 Sydney 2012. The index case on October 31 was followed by 4 waves of secondary and tertiary cases. Residents were the most affected group in this outbreak, with a high incidence rate, which is common in AGE outbreaks affecting this particular population.Reference Fernández, Truyols, Guibert, Cerdá and Gayá 18 NoV infections can cause severe disease in the elderly, including prolonged symptoms and death.Reference Chen, Hall, Kirk, Health and Diseases 2 , Reference Beersma, Sukhrie and Bogerman 4
All confirmed cases carried the emergent NoV GII.P16-GII.4 Sydney 2012 with 100% nucleotide sequence homology, suggesting a common-source outbreak. This is the first report of the circulation of this lineage in Portugal. Nonetheless, it has been reported in several countries including the United States, Japan, Germany, France, England, and Brazil since late 2014.Reference Cannon, Barclay and Collins 19 – Reference Barreira, Fumian and Tonini 24
The 4 consecutive peaks in the epidemic curve, the clustering of cases in their working or living areas, and the highest incidence rate in nurses compared to other staff members point to person-to-person transmission. This finding is consistent with previous studies reporting person-to-person transmission as the main route of NoV transmission in LTCFs.Reference Chen, Hall, Kirk, Health and Diseases 2 , Reference Cardemil, Parashar and Hall 5 , Reference Cui, Pan and Wang 25 This transmission was likely due to the caregiving and close contact required between staff and residents with limited mobility and because institutionalized confinement promotes transmission by sharing rooms and touching common surfaces. Therefore, control of NoV outbreaks is extremely difficult in healthcare facilities, as evidenced in this outbreak, which lasted 1 month and affected 146 people.
This outbreak had a delay in reporting to the local public health authority, a delay in the identification of causative agent, and a lack of strict control measures. For instance, a staffing shortage due to the exclusion from work of cases among staff members in the beginning of the outbreak led to the need for all categories of staff members to move, which facilitated the transmission between wards. Control measures were only effectively implemented from November 25 onward, when isolation and cohorting of cases was fully implemented with no exceptions. The restrictions imposed by the local public health authorities apparently contributed to the outbreak control among staff members on November 26. However, a new wave of transmission occurred among residents after that time, starting in the ward where the last cases had been observed among staff members. More cases appeared in another 6 wards during the following 2 weeks. Symptomatic patients and healthcare workers (HCWs) are more often involved in transmission events. Nevertheless, this transmission could be explained by asymptomatic shedders (HCWs and patients), which has been described previously.Reference Teunis, Sukhrie, Vennema, Bogerman, Beersma and Koopmans 26 , Reference Miura, Matsuyama and Nishiura 27
The study had several limitations. First, the notification of a suspected AGE outbreak was only sent to the local public health unit 9 days after the occurrence of the first cases. The focus was to control the outbreak as soon as possible, but an investigation to identify the potential source was not conducted. Secondly, the retrospective epidemiological questionnaires completed by staff members did not provide additional information compared to the questionnaires completed during the outbreak, which may have been due to recall bias because time had elapsed since the outbreak. Nevertheless, the high incidence rates, knowledge of the movement of residents and staff members between wards, and the genotype assessment pointed toward the emergent GII.P16-GII.4 Sydney 2012 NoV being transmitted person to person as the most likely cause of the outbreak. The outbreak ended only after the implementation of strict control measures, suggesting that these current recommendations for controlling NoV outbreaks in LTCFs must be rigorously followed.
Acknowledgments
We acknowledge the facilitating role and the active measures implemented by Ana Reis, the clinical director, Célia Pereira, the director nurse, and Guilherme Rodrigues, the nurse responsible of the local control group for infection control. In addition, this material is based upon work supported by the fellowship European Public Health Microbiology Training (EUPHEM), European Centre for Disease Prevention and Control (ECDC).
Financial support
No financial support was provided relevant to this article.
Conflicts of interest
All authors report no conflicts of interest relevant to this article.






